Biophysics.

Researchers in the Biophysics area at BIFI apply experimental and computational tools and methodologies in an interdisciplinary environment for understanding the behavior of biological systems, from molecules (proteins, nucleic acids, small molecules, etc.) to whole organisms and ecosystems, from a quantitative perspective. Proteins and organisms with biotechnological and/or biomedical relevance are of special interest. Research within this area has many applications in different fields:
Biotechnology (protein stabilization and modeling, protein function design, molecular machines, nanostructures, molecular modeling, electronic transport, catalysis, electromagnetic interaction with matter, and protein glycosylation and its role in signaling)
Biomedicine (drug discovery and design, pharmacological target identification and validation, protein-DNA interaction)
Biology (epidemiology, evolution, complex networks)
The Biophysics area at BIFI encompasses 9 lines of research:
- Protein folding and molecular design
- Biomolecular interactions
- Protein glycosylation and its role in disease
- Flavoenzymes: action mechanisms and biotecnology
- Protein misfolding and amyloid aggregation
- Clinical diagnosis and drug delivery
- Signal transduction and membrane protein therapies
- Enzyme modulation & reaction mechanisms
- Structural Biology of neuronal membrane receptors
- Protein folding and molecular design
- Biomolecular Interactions
- Protein glycosylation and its role in disease
- Flavoenzymes: action mechanisms and biotecnology)
- Protein misfolding and amyloid aggregation
- Clinical diagnosis and drug delivery
- Structural Biology of neuronal membrane receptors
- Signal transduction and membrane protein therapies
- Enzyme modulation & Reaction Mechanisms
Protein folding and molecular design
Head of the Research Line:
Javier Sancho
Researchers:
Dr. Juan José Galano Frutos
Dr. Helena García Cebollada
Ritwik Maity
Patricia Bruñen Fau
Dr. Verónica Iguarbe Montalbán
Darío Bazco Marco
Antonio Hidalgo Toledo
Alfonso López
SUMMARY OF THE RESEARCH LINE
 We seek a quantitative understanding of proteins thermodynamics that allows us to govern the composition of protein ensembles. Using a variety of biophysical techniques, we determine the essential thermodynamic magnitudes of the protein folding equilibrium (ΔG, ΔH, ΔS and ΔCp) in wild type proteins and in variants obtained using protein engineering techniques.
We seek a quantitative understanding of proteins thermodynamics that allows us to govern the composition of protein ensembles. Using a variety of biophysical techniques, we determine the essential thermodynamic magnitudes of the protein folding equilibrium (ΔG, ΔH, ΔS and ΔCp) in wild type proteins and in variants obtained using protein engineering techniques.
We analyse Molecular Dynamics simulations to increase our understanding of the basic principles. We try to understand the contribution of fundamental interactions (e.g. different types of electrostatic interactions) but also of chemical entities (side chains of amino acid residues) to protein stability. We aspire to provide conceptual and computational tools to make possible a truly quantitative design of protein stability.
HTS, ANTIMICROBIALS, PHARMACOLOGICAL CHAPERONES, MEDICINAL CHEMISTRY
 We use our knowledge to develop drugs that target proteins, either human or otherwise. We can start from a defined protein target, identify hits, and develop them into leads using a full medicinal chemistry approach.
We use our knowledge to develop drugs that target proteins, either human or otherwise. We can start from a defined protein target, identify hits, and develop them into leads using a full medicinal chemistry approach.
Our more advanced projects relate to novel bug-specific antimicrobials against Helicobacter pylori, personalized pharmacological chaperons for phenylketonuria, and novel aggregation inhibitors for Parkinson disease.
BIOINFORMATICS, PREDICTORS, MD SIMULATIONS, HUMAN VARIOME
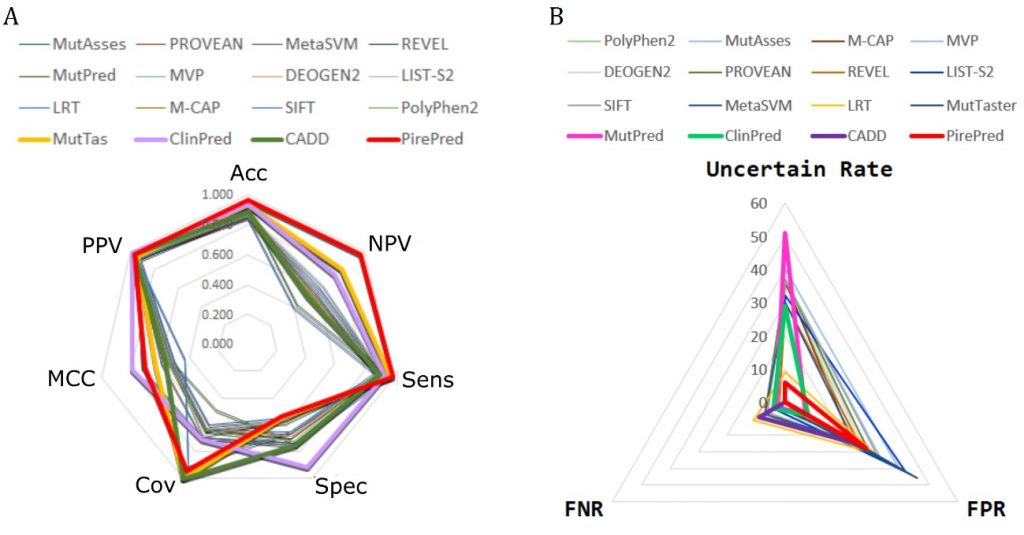
We are developing techniques to increase the accuracy of phenotypic interpretation of single nucleotide genetic variants. We try to boost the use of MD techniques to accurately classify genetic variants into benign and deleterious. We focus in missense variants affecting regions coding for proteins with a well defined function.
Among others, we work in the interpretation of variants in genes related to diseases screened in the newborn.
Relevant publications
1. Galano-Frutos, Juan José; Nerín-Fonz, Francho; Sancho, Javier. 2023. Calculation of Protein Folding Thermodynamics using Molecular Dynamics Simulations. J. Chem. Inf. Model. In Press
2. Galano-Frutos, Juan José, Torreblanca Renzo, García-Cebollada Helena, Sancho Javier. 2022. A look at the face of the molten globule: structural model of the Helicobacter pylori apoflavodoxin ensemble at acidic pH. Protein Science 2022,31:e4445
3. Galano-Frutos, Juan José; García-Cebollada, Helena; Sancho, Javier. 2021. Molecular Dynamics Simulations for Genetic Interpretation in Protein Coding Regions: Where we Are, Where to Go and When. Briefings in bioinformatics. 22:3–192.
4. Pujols, J.; et al. (18/14) 2018. Small molecule inhibits alpha-synuclein aggregation, disrupts amyloid fibrils, and prevents degeneration of dopaminergic neurons. Proceedings of the national academy of sciences. 115-41, pp.10481-10486. ISSN 0027-8424.
5. Espinosa Angarica, Vladimir; Orozco, Modesto; Sancho, Javier. 2016. Exploring the complete mutational space of the LDL receptor LA5 domain using molecular dynamics: Linking snps with disease phenotypes in familial hypercholesterolemia. Human molecular genetics. 25-6, pp.1233-1246. ISSN 0964-6906.
Main research projects
1. MOlecular-Scale Biophysics Research Infrastructure MOSBRI. UE (H2020-INFRAIA-2020-1). 2021-2025. IP: Javier Sancho (Coordinator: Patrick England, Institute Pasteur, France)
2. Estabilizador automático de proteínas para biotecnología y biomedicina. Ministerio de Ciencia e Innovación. 2021-2024. IP: Javier Sancho
3. La estabilidad de proteínas: análisis en profundidad, nuevas herramientas de predicción y aplicaciones en biomedicina y biotecnología. PID2022-141068NB-I00. Ministerio de Ciencia e Innovación. 2023-2026. IP: Javier Sancho
Collaborators
We maintain active collaborations with many national and international groups and networks: INPEC. See publications. See also project webs: RedMut, Pirepred.
CONTACT
Biomolecular Interactions
Head of the Research Line:
Adrian Velazquez-Campoy
Researchers:
José Luis Neira (Universidad Miguel Hernández, Elche)
Bruno Rizzuti (Institute of Nanotechnology, CNR, Italia)
David Ortega Alarcón
Ana Jiménez Alesanco
Paula García Franco
Ana Jiménez Alesanco
SUMMARY OF THE RESEARCH LINE
All biological processes can be described as a sequence of interaction processes between biomolecules and coupled conformational changes. We work on three fundamental questions, from both a basic and applied perspective, related to interactions in biological molecules:
- Biophysical characterization of proteins of biotechnological and/or biomedical relevance: conformational and functional landscape, interactions with other biomolecules, conformational changes, phase diagrams, cooperative phenomena….
- Development of experimental methodologies for the study of biomolecular interactions, cooperative phenomena and functional regulation in biological systems.
- Development and implementation of high-throughput screening procedures for the identification of bioactive compounds capable of modulating the function of pharmacological protein targets.
Relevant publications
1. N. Losada-Garcia, A. Vazquez-Calvo, D. Ortega-Alarcon, O. Abian, A. Velazquez-Campoy, P. Domingo-Calap, A. Alcami, J.M. Palomo. Nanostructured biohybrid material with wide-ranging antiviral action. Nano Research 2023, 16:11455-11463
2.- S. Araujo-Abad, B. Rizzuti, A. Villamarin-Ortiz, D. Pantoja-Uceda, C.M. Moreno-Gonzalez, O. Abian, A. Velazquez-Campoy, J.L. Neira, C. de Juan Romero. New insights into cancer: MDM2 binds to the citrullinating enzyme PADI4. Protein Science 2023, 32:e4723
3.– S. Araujo-Abad, M. Fuentes-Baile, B. Rizzuti, J.F. Bazan, A. Villamarin-Ortiz, M. Saceda, E. Fernandez, M. Vidal, O. Abian, A. Velazquez-Campoy, C. de Juan Romero, J.L. Neira. The intrinsically disordered, epigenetic factor RYBP binds to the citrullinating enzyme PADI4 in cancer cells. International Journal of Biological Macromolecules 2023, 246:125632
4.– M. Bastos, O. Abian, C.M. Johnson, F. Ferreira-da-Silva, S. Vega, A. Jimenez-Alesanco, D. Ortega-Alarcon, A. Velazquez-Campoy. Isothermal titration calorimetry. Nature Reviews Methods Primers 2023, 3:17
5.– D. Ortega-Alarcon, R. Claveria-Gimeno, S. Vega, O.C. Jorge-Torres, M. Esteller, O. Abian, A. Velazquez-Campoy. Unexpected thermodynamic signature for the interaction of hydroxymethylated DNA with MeCP2. International Journal of Biological Macromolecules 2023, 232:123373
6.– K. Gonzalez-Arzola, A. Diaz-Quintana, N. Bernardo-Garcia, J. Martinez-Fabregas, F. Rivero-Rodriguez, M.A. Casado-Combreras, C. Elena-Real, A. Velazquez-Cruz, S. Gil-Caballero, A. Velazquez-Campoy, E. Szulc, M.P. Gavilan, I. Ayala, R. Arranz, R.M. Rios, X. Salvatella, J.M. Valpuesta, J. Hermoso, M.A. De la Rosa, I. Diaz-Moreno. Nucleus-translocated mitochondrial cytochrome c liberates nucleophosmin-sequestered ARF tumor suppressor by changing nucleolar liquid-liquid phase separation. Nature Structural & Molecular Biology 2022, 29:1024-1036
7.– A. Jimenez-Alesanco, U. Eckhard, M. Asencio del Rio, S. Vega, T. Guevara, A. Velazquez-Campoy, F.X. Gomis-Rüth, O. Abian. Repositioning small molecule drugs as allosteric inhibitors of the BFT-3 toxin from enterotoxigenic Bacteroides fragilis. Protein Science 2022, 31:e4427
8.– D. Ortega-Alarcon, R. Claveria-Gimeno, S. Vega, O.C. Jorge-Torres, M. Esteller, O. Abian, A. Velazquez-Campoy. Stabilization effect of intrinsically disordered regions on multidomain proteins: The case of the methyl-CpG protein 2, MeCP2. Biomolecules 2021, 11:1216
9.– D. Ortega-Alarcon, R. Claveria-Gimeno, S. Vega, O.C. Jorge-Torres, M. Esteller, O. Abian, A. Velazquez-Campoy. Influence of the disordered domain structure of MeCP2 on its structural stability and dsDNA interaction. International Journal of Biological Macromolecules 2021, 175:58-66
10.– B. Rizzuti, L. Ceballos-Laita, D. Ortega-Alarcon, A. Jimenez-Alesanco, S. Vega, F. Grande, F. Conforti, O. Abian, A. Velazquez-Campoy. Sub-micromolar inhibition of SARS-CoV-2 3CLpro by natural compounds. Pharmaceuticals 2021, 14:892
11. O. Abian, D. Ortega-Alarcon, A. Jimenez-Alesanco, L. Ceballos-Laita, S. Vega, H.T. Reyburn, B. Rizzuti, A. Velazquez-Campoy. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. International Journal of Biological Macromolecules 2020, 164:1693-1703
12.– J. Felix, K. Weinhäupl, C. Chipot, F. Dehez, A. Hessel, D.F. Gauto, C. Morlot, O. Abian, I. Gutsche, A. Velazquez-Campoy, P. Schanda, H. Fraga. Mechanism of the allosteric activation of the ClpP protease machinery by substrates and active-site inhibitors. Science Advances 2019, 5:eaaw3818
13.– P. SaNtofimia‐Castaño, Y. Xia, W. Lan, Z. Zhou, C. Huang, L. Peng, P. Soubeyran, A. Velazquez‐Campoy, O. Abian, B. Rizzuti, J.L. Neira, J. Iovanna. Ligand‐based design of a potent inhibitor of NUPR1 exerting anticancer activity via necroptosis. Journal of Clinical Investigation 2019, 129:2500-2513
14.– J.L. Neira, J. Bintz, M. Arruebo, B. Rizzuti, T. Bonacci, S. Vega, A. Lanas, A. Velazquez-Campoy, J.L. Iovanna, O. Abian. Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Scientific Reports 2017 7:39732
15.– R. Claveria-Gimeno, P.M. Lanuza, I. Morales-Chueca, O.C. Jorge, S. Vega, O. Abian, M. Esteller, A. Velazquez-Campoy. The intervening domain from MeCP2 enhances the DNA affinity of the methyl binding domain and provides an independent DNA interaction site. Scientific Reports 2017 7:41635
Main research projects
1. New pharmacological strategy against zinc-dependent target proteins: Allosteric inhibitors of deacetylases HDAC8 and LpxC. Ministerio de Ciencia e Innovación (PID2021-127296OB-I00). Entidades participantes: Universidad de Zaragoza. 2022-2025. IP: Adrián Velázquez Campoy (Universidad de Zaragoza – BIFI)
2.Nuevas estrategias contra el cáncer: Inhibición de las interacciones moleculares de las proteínas deiminasas de arginina (InterPATh). Generalitat Valenciana (CIAICO/2021/135). 2022-2024. Entidades participantes: Universidad Miguel Hernández. IP: Meuri del Camino de Juan Romero – José L. Neira (Universidad Miguel Hernández
3.MOlecular-Scale Biophysics Research Infrastructure (MOSBRI). Horizon 2020 – Research and Innovation Framework Programme (H2020-INFRAIA-2020-1, 101004806, RIA). Entidades participantes: Institut Pasteur, Aarhus Universitet, Universite Libre de Bruxelles, Universita Degli Studi di Genova, Rijksuniversiteit Groningen, European Molecular Biology Laboratory, Linkopings Universitet, Kemijski Institut, Birkbeck College – University of London, Centre National de la Recherche Scientifique, Biotechnologicky Ustav AV CR, Universita degli Studi di Roma La Sapienza, Universidad de Zaragoza, Malvern Panalytical, Software 4 Science Developments. 2021-2025. IP: Patrick England (Main Proposer, Institut Pasteur), Adrian Velazquez Campoy (Proposer, Universidad de Zaragoza – BIFI)
Collaborators
- Olga Abian (Universidad de Zaragoza y BIFI, Spain)
- Juan Ausio (University of Victoria, Canada)
- Margarida Bastos (University of Porto, Portugal)
- Pierpaolo Bruscolini (Universidad de Zaragoza y BIFI, Spain)
- Jose A. Carrodeguas (Universidad de Zaragoza y BIFI, Spain)
- Irene Diaz-Moreno (Instituto de Bioquímica Vegetal y Fotosíntesis – CSIC, Spain)
- Manel Esteller (Instituto de Investigación contra la Leucemia Josep Carreras, Spain)
- Maria Fillat (Universidad de Zaragoza y BIFI, Spain)
- Hugo Fraga (Instituto de Investigacao e Inovacao em Saude, Portugal)
- Ernesto Freire (The Johns Hopkins University, USA)
- Sònia Guil (Instituto de Investigación contra la Leucemia Josep Carreras, Spain)
- Ruben Martinez-Buey (Universidad de Salamanca, Spain)
- Milagros Medina (Universidad de Zaragoza y BIFI, Spain)
- Maria João Moreno (Universidade de Coimbra, Portugal)
- Arturo Muga (Universidad del Pais Vasco, Spain)
- Santiago Ramon-Maiques (CNIO, Spain)
- David Reverter (Universidad Autonoma de Barcelona, Spain)
- Javier Sancho (Universidad de Zaragoza y BIFI, Spain)
- Jayaraman Sivaraman (National University of Singapore, Singapore)
- Salvador Ventura (Universidad Autonoma de Barcelona, Spain)
CONTACT
Adrian Velazquez-Campoy, adrianvc@unizar.es
Protein glycosylation and its role in disease
Head of the Research Line:
Ramón Hurtado Guerrero
Researchers:
Víctor Taleb
Billy Veloz
David Sánchez
Irene Ginés
María Bort
SUMMARY OF THE RESEARCH LINE
Our group is interested in the study of glycosyltransferases, glycosylhydrolases and carbohydrate-binding proteins/modules involved in human diseases. We use protein X-ray crystallography as the main tool complemented with enzymology, inhibition studies, etc, in order to study the molecular mechanisms of enzymes that are involved in the synthesis, modification and degradation of glycoconjugates, oligo and polysaccharides (see more relevant publications below).
We majorly work in Protein O-glycosylation and glycosyltransferases responsible of this post-translational modification. In particular we are currently working in several glycosyltransferases such as Protein O-fucosyltransferases 1 and 2 (POFUT1 and 2), FUT8 and the large family of GalNAc-Ts. While POFUT1 and 2 fucosylate folded domains such as EFG and TSR domains, respectively, GalNAc-Ts mainly glycosylate unstructured regions present in a large number of proteins such as mucins . FUT8 is responsible of the so-called core fucosylation (Figure 1).
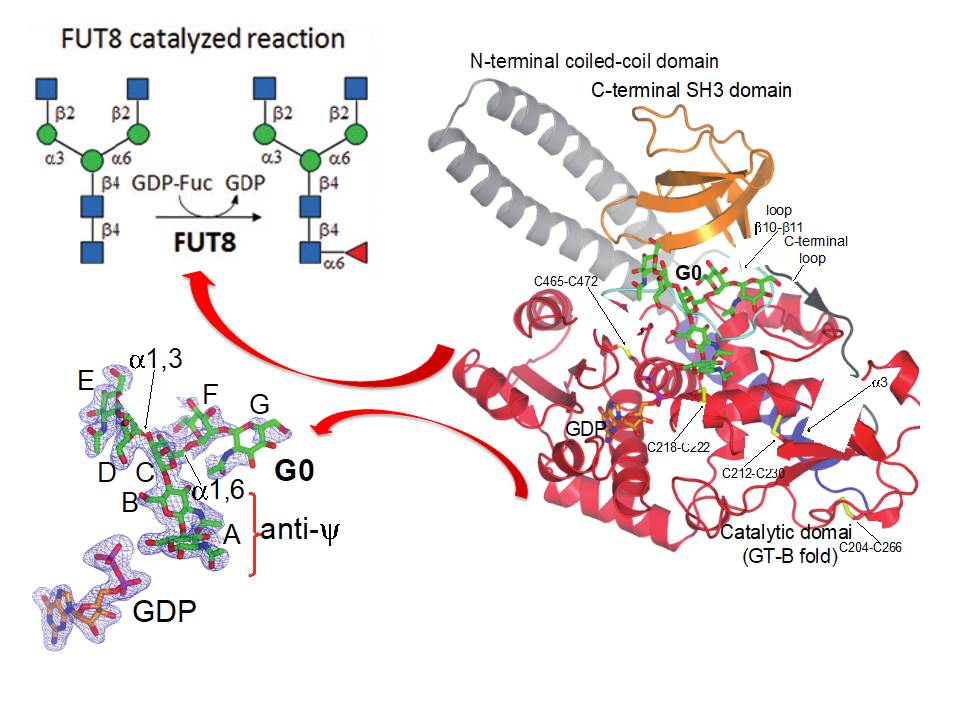
Figure 1. Scheme summarizing the structure of FUT8 complexed to GDP and a biantennary N-glycan.
Furthermore we are interested in the elucidation of the reaction coordinates and the molecular mechanism by using transition state analogues or Michaelis complexes. Finally these studies will be important for the design of new future drugs with potential therapeutic applications.
Although not related to the Glycobiology field, we have a close collaboration with Prof. Guadix and Dr Conejo-García from University of Granada in the development of new inhibitors against human choline kinase α1.
Relevant publications:
10 selected publications since 2013, *corresponding author(s)
1. Substrate-guided front-face reaction revealed by combined structural snapshots and metadynamics for the polypeptide GalNAc-T2. Lira-Navarrete E, Iglesias-Fernández J, Zandberg WF, Compañón I, Kong Y, Corzana F, Pinto BM, Clausen H, Peregrina JM, Vocadlo DJ, Rovira C*, Hurtado-Guerrero R*. (2014) Angew Chem Int Ed Engl 53(31):8206-10. (89 citations/9 yrs)
First trapped experimental Michaelis complex for a retaining glycosyltransferase probing the SNi mechanism
2. Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Lira-Navarrete E, de Las Rivas M, Compañón I, Pallarés MC, Kong Y, Iglesias-Fernández J, Bernardes GJ, Peregrina JM, Rovira C, Bernadó P, Bruscolini P, Clausen H, Lostao A, Corzana F, Hurtado-Guerrero R*. (2015). Nat Commun 6:6937. (92 citations/8 yrs)
First work describing how the lectin domain of GalNAc-Ts modulates O-glycosylation
3. A proactive role of water molecules in acceptor recognition by protein O-fucosyltransferase 2. Valero-González J, Leonhard-Melief C, Lira-Navarrete E, Jiménez-Osés G, Hernández-Ruiz C, Pallarés MC, Yruela I, Vasudevan D, Lostao A, Corzana F, Takeuchi H, Haltiwanger RS, Hurtado-Guerrero R*. (2016) Nat Chem Biol 12(4):240-6. (65 citations/7 yrs)
Describes the key role of water molecules in the binding of PoFUT2 to its various protein substrates
4. The interdomain flexible linker of the polypeptide GalNAc transferases dictates their long-range glycosylation preferences. de Las Rivas M, Lira-Navarrete E, Daniel EJP, Compañón I, Coelho H, Diniz A, Jiménez-Barbero J, Peregrina JM, Clausen H, Corzana F, Marcelo F, Jiménez-Osés G, Gerken TA, Hurtado-Guerrero R*. (2017) Nat Commun 8(1):1959. (35 citations/6 yrs)
First elucidation of the role of a small flexible linker dictating the long-glycosylation preference
5. Structural basis for arginine glycosylation of host substrates by bacterial effector proteins. Park JB, Kim YH, Yoo Y, Kim J, Jun SH, Cho JW, El Qaidi S, Walpole S, Monaco S, García-García AA, Wu M, Hays MP, Hurtado-Guerrero R*, Angulo J*, Hardwidge PR, Shin JS*, Cho HS*. (2018) Nat Commun 9(1):4283. (52 citations/5 yrs)
Elucidation of how arginine-glycosyltransferases from entheropathogens glycosylate human proteins to regulate immunological activity
6. Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases. Fang W, Sanz AB, Bartual SG, Wang B, Ferenbach AT, Farkaš V, Hurtado-Guerrero R, Arroyo J*, van Aalten DMF*. (2019). Nat Commun 10(1):1669. (21 citations/4 yrs)
Detailed study of the importance of the Crh transglycosylases in the biology of A. fumigatus
7. Structural basis for substrate specificity and catalysis of α1,6-fucosyltransferase García-García A, Ceballos-Laita L, Serna S, Artschwager R, Reichardt NC, Corzana F, Hurtado-Guerrero R*. (2020). Nat Commun 2020;11(1):973. (37 citations/3 yrs)
Structural basis of how the fucosyltransferase FUT8 achieves core-fucosylation on N-glycans
8- Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3. de Las Rivas M, Paul Daniel EJ, Narimatsu Y, Compañón I, Kato K, Hermosilla P, Thureau A, Ceballos-Laita L, Coelho H, Bernadó P, Marcelo F, Hansen L, Maeda R, Lostao A, Corzana F, Clausen H, Gerken TA, Hurtado-Guerrero R*. (2020). Nat Chem Biol 16(3):351-360. (42 citations/3 yrs)
Describes the molecular and cellular mechanisms underlying the regulation of FGF23 by GalNAc-T3
9. Structural basis for the synthesis of the core 1 structure by C1GalT1. González-Ramírez AM, Grosso AS, Yang Z, Compañón I, Coelho H, Narimatsu Y, Clausen H, Marcelo F, Corzana F*, Hurtado-Guerrero R*. (2022) Nat Commun 13(1):2398. (6 citations/1 yr)
Molecular insights into the synthesis of core1 by C1GalT1
10. Structural and mechanistic insights into the cleavage of clustered O-glycan patches-containing glycoproteins by mucinases of the human gut. Taleb V, Liao Q, Narimatsu Y, García-García A, Compañón I, Borges RJ, González-Ramírez AM, Corzana F, Clausen H, Rovira C*, Hurtado-Guerrero R*. (2022) Nat Commun 13(1):4324. (7 citations/1 yr)
Molecular insights into AM0627 function from X-ray crystallography and computer simulations
11. Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase. Piniello B, Macías-León J, Miyazaki S, García-García A, Compañón I, Ghirardello M, Taleb V, Veloz B, Corzana F, Miyagawa A, Rovira C*, Hurtado-Guerrero R*. (2023) Nat Commun 14(1):5785.
Structural and Molecular insights into the N-glycosylation by a soluble N-glycosyltransferase
12. Identification of global inhibitors of cellular glycosylation. Sørensen DM, Büll C, Madsen TD, Lira-Navarrete E, Clausen TM, Clark AE, Garretson AF, Karlsson R, Pijnenborg JFA, Yin X, Miller RL, Chanda SK, Boltje TJ, Schjoldager KT, Vakhrushev SY, Halim A, Esko JD, Carlin AF, Hurtado-Guerrero R, Weigert R, Clausen H, Narimatsu Y. (2023) . Nat Commun. 14(1):948.
Discovery of inhibitors affecting several pathways of glycosylation.
Current Main research projects
1. PID2019-105451GB-I00.(AEI, Spain), 2020-2023. 266,200 euros plus extra funding for one PhD student. PI: Ramón Hurtado-Guerrero
2. Desarrollo de nuevas inmunoterapias para el tratamiento del cáncer y candidiasis. LMP10_21 (2021-2023). DGA. PI: Ramón Hurtado-Guerrero. 54,500 euros.
3. Nuevos tratamientos de inmunoterapia (Nanobodies y células CAR) frente a infecciones fúngicas invasivas en pacientes oncopediátricos. Convocatoria de Ayudas a la investigación del cáncer infantil. Aspanoa. 2022-2023. IP: Eva Gálvez and Maykel Arias. coIPs: Ramón Hurtado-Guerrero and Julián Pardo. 60.000 euros
4. Targeting GALNT7 to develop new drugs for the personalised treatment of prostate cancer. Prostate Cancer UK. PIs: Jennifer Munkley and Ben Schumann. Collaborators: Ramón Hurtado-Guerrero and others. 2022-2025.
5. GlyCanDrug. European Commission (ITN), HORIZON-MSCA-2022-DN-01. 251,971.20 to my group, 2024-2027. Total to the ITN = 2,696,457.60
6. PID2022-136362NB-I00 (AEI, Spain), 2023-2026. 375,000 euros plus extra funding for one PhD student. PI: Ramón Hurtado-Guerrero
Collaborators
Robert Haltiwanger, The University of Georgia
Henrik Clausen, University of Copenhagen
Daan van Aalten, University of Dundee
Philip Hardwidge, Kansas State University
Tom Gerken, Case Western Reserver University
Pedro Merino, Universidad de Zaragoza
Francisco Corzana, Universidad de La Rioja
Carme Rovira, Universidad de Barcelona
Filipa Marcelo, New University of Lisbon
Julián Pardo, Universidad de Zaragoza
Anabel Lostao, Universidad de Zaragoza
Flavoenzymes: action mechanisms and biotecnology
Head of the Research Line:
Dr. Patricia Ferreira Neila
Dr. Marta Martínez Júlvez
Researchers:
Dr. Milagros Medina Trullenque
Dr. Patricia Ferreira Neila
Dr. Marta Martínez Júlvez
Sergio Boneta Martinez
Andrea Moreno Maldonado
Olga Arjona Soriano
Maribel Bernabé Rivero
Diego Boj Carballo
Paula Cinca Fernando
Víctor Correa Pérez
Miguel Ferrer Navarro
Aurora Victoria Vázquez Rodríguez
SUMMARY OF THE RESEARCH LINE
Flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) cofactors enable flavoproteins and flavoenzymes to exhibit significant redox versatility in a wide range of physiological catalytic processes, primary and secondary metabolisms, as well as interactions with other biomolecules. Furthermore, among the approximately 400 different flavoproteins identified, nearly 10% act in non-redox reactions or as signaling and sensing molecules. Some may act as suitable biocatalysts due to the selectivity, control, and efficiency in the reactions they engage in, while others may serve as therapeutic targets in the treatment of infectious diseases and mammalian pathological disorders. However, their extensive diversity, species-specific features, as well as interplay with themselves, other proteins and metabolites, mean that we are still far from understanding how to exploit their full potential.
In this context, the main goal of our team is to provide better comprehension of molecular mechanisms for key metabolic enzymes, with particular emphasis on flavoenzyme-dependent systems. This will help clarify their roles in the metabolic processes and interacting networks in which they participate, as well as to establish the molecular bases to facilitate their applicability as therapeutic targets or biocatalysts.
Our main research topics include:
- Unraveling of the molecular bases that determine the cellular functions of human flavoenzymes involved in cell bioenergetics, signaling or apoptosis in both healthy and pathologic scenarios. This knowledge will pave for the development of novel therapeutic strategies to overcome the effects of pathogenic mutations.
- Discovering the flavoproteome content in relevant species and investigating the action mechanism of their flavoprotein and flavoenzyme systems. This exploration aims to enable their use as targets in the search for antimicrobials and/or new biotechnological tools.
- Understanding the mechanisms for flavin incorporation into client apo-proteins and the impact of flavin endogenous modifications on flavoprotein functionality.
- Developing a novel catalog of biocatalysts for the application in sustainable synthetic chemistry towards the production of bio-based compounds of interest.
- Unravel the contribution of binding and dynamics processes during the action mechanisms of the studied systems through a combination of structural computational and experimental approaches.
In our research, we combine a large number of biochemical, biophysical, molecular biology and structural biology tools together computational approaches for the expression, purification, rational design and structural-functional characterization of proteins and their interacting networks.
The main methodologies used in our research include:
- Production of native and mutant proteins through protein engineering techniques.
- Homologous and heterologous protein expression in different microorganisms.
- Purification of proteins and separation of metabolites (electrophoresis, chromatographic methods by FPLC and HPLC, …).
- Absorption spectrometry: kinetic studies in steady-state, differential spectroscopy, binding affinity, midpoint reduction potentials determination.
- Transient kinetics by multimixing stopped-flow spectroscopy.
- Interaction and kinetic assays under anaerobic conditions.
- Fluorescence spectroscopy and circular dichroism.
- Protein crystallization, x-ray diffraction and starting time-resolved serial femtosecond crystallography (SFX) and X-ray Free-Electron Lasers (XFELs).
- Advanced Electron Paramagnetic Resonance related techniques (ESEEM, HYSCORE, ENDOR).
- Isothermal Titration Calorimetry and Differential Scanning Calorimetry.
- Computational Biology Methods: Docking, Molecular Dynamics and QM/MM simulations.
- Bioinformatic tools for protein target identification and selection
- High-throughput screening of chemical libraries of compounds.
RELEVANT PUBLICATIONS
1. Increased demand for FAD synthesis in differentiated and stem pancreatic cancer cells is accomplished by modulating FLAD1 gene expression: the inhibitory effect of Chicago Sky Blue. Nisco A, Carvalho TMA, Tolomeo M, Di Molfetta D, Leone P, Galluccio M, Medina M, Indiveri C, Reshkin SJ, Cardone RA, Barile M. FEBS J. 2023 Oct;290(19):4679-4694. doi: 10.1111/febs.16881
2. Beyond a platform protein for the degradosome assembly: The Apoptosis-Inducing Factor as an efficient nuclease involved in chromatinolysis. 2023. Novo N, Romero-Tamayo S, Marcuello C, Boneta S, Blasco-Machin I, Velázquez-Campoy A, Villanueva R, Moreno-Loshuertos R, Lostao A, Medina M, Ferreira P. PNAS Nexus. 2022 26;2(2):pgac312. doi: 10.1093/pnasnexus/pgac312. eCollection 2023 Feb.
3. Riboflavin kinase and pyridoxine 5′-phosphate oxidase complex formation envisages transient interactions for FMN cofactor delivery. 2023. Rivero M, Boneta S, Novo N, Velázquez-Campoy A, Polo V, Medina M. Front Mol Biosci. 10:1167348. doi: 10.3389/fmolb.2023.1167348. eCollection 2023.
4. Expanding the Physiological Role of Aryl-Alcohol Flavooxidases as Quinone Reductases. 2023. Ferreira P, Carro J. Balcells B, Martinez AT, Serrano A. Applied and Environmental Microbiology. May 31;89(5):e0184422. doi: 10.1128/aem.01844-22.
5. Counterintuitive structural and functional effects due to naturally occurring mutations targeting the active site of the disease-associated NQO1 enzyme. 2023. Pacheco-García JL, Anoz-Carbonell E, Loginov DS, Kavan D, Salido E, Man P, Medina M, Pey AL. FEBS J. 2023 Apr;290(7):1855-1873. doi: 10.1111/febs.16677.
6. A global phylogenomic analysis of the shiitake genus Lentinula. Sierra-Patev S, Byoungnam M, Naranjo-Ortiz M, Looney B, Konkel Z, Slot JC, Sakamoto Y, Steenwyk JL, Rokas A, Carro J, Camarero S, Ferreira P, … Hibbett D. 2023. PNAS. 120(10):e2214076120. doi: 10.1073/pnas.2214076120.
7. Mining the Flavoproteome of Brucella ovis, the Brucellosis: Causing Agent in Ovis aries. 2022. Minjárez-Sáenz M, Martínez-Júlvez M, Yruela I, Medina M. Microbiol Spectr. 10(2):e0229421. doi: 10.1128/spectrum.02294-21.
8. Cofactors and pathogens: FMN and FAD biosynthesis by the FAD synthase from Brucella ovis. 2022. Moreno SA, Taleb V, Sebastián M, Anoz-Carbonell EA, Martínez-Júlvez M, Medina M. IUBMB LIFE. 74:655-671. doi: 10.1002/iub.2576.
9. Atomic Force Microscopy to Elicit Conformational Transitions of Ferredoxin-Dependent Flavin Thioredoxin Reductases. 2021. Marcuello C, Frempong GA, Balsera M, Medina M, Lostao A. Antioxidants 9;10(9):1437. doi: 10.3390/antiox10091437.
10. Structural basis of the pleiotropic and specific phenotypic consequences of missense mutations in the multifunctional NAD(P)H:quinone oxidoreductase 1 and their pharmacological rescue. 2021. Pacheco-Garcia JL, Anoz-Carbonell E, Vankova P, Kannan A, Palomino-Morales R, Mesa-Torres N, Salido E, Man P, Medina M, Naganathan AN, Pey AL. Redox Biol. 46:102112. doi: 10.1016/j.redox.2021.102112.
11. W196 and the β-Hairpin Motif Modulate the Redox Switch of conformation and the Biomolecular Interaction Network of the Apoptosis-Inducing Factor. 2021. Romero-Tamayo S, Laplaza R, Velázquez-Campoy A, Villanueva R, Medina M, P. Ferreira P. Oxid Med Cell Longev. 15;2021:6673661. doi: 10.1155/2021/6673661. eCollection 2021.
12. Unexpected diversity of ferredoxin-dependent thioredoxin reductases in cyanobacteria. Buey RM, Fernández-Justel D, González-Holgado G, Martínez-Júlvez M, González-López A, Velázquez-Campoy A, Medina M, Buchanan BB, Balsera M. Plant Physiol. 2021 May 27;186(1):285-296. doi: 10.1093/plphys/kiab072.
13. Anaerobic Stopped-Flow Spectrophotometry with Photodiode Array Detection in the Presteady State: An Application to Elucidate Oxidoreduction Mechanisms in Flavoproteins Ferreira P.; Medina M. 2021. METHODS IN MOLECULAR BIOLOGY. 2280: 135-155. doi: 10.1007/978-1-0716-1286-6_9
14. The Catalytic Cycle of the Antioxidant and Cancer-Associated Human NQO1 Enzyme: Hydride Transfer, Conformational Dynamics and Functional Cooperativity. Anoz-Carbonell E, Timson DJ, Pey AL, Medina M. Antioxidants (Basel). 2020 Aug 20;9(9):772. doi: 10.3390/antiox9090772
15. Redox-and ligand binding-dependent conformational ensembles in the human apoptosis-inducing factor regulate its pro-life and cell death functions. Villanueva R.; Romero-Tamayo S.; Laplaza R.; Martinez-Olivan J.; Velazquez-Campoy A.; Sancho J.; Ferreira P*; Medina M*.. 2019. ANTIOXIDANTS & REDOX SIGNALING. 30 (18): 2013-2029. doi: 10.1089/ars.2018.7658.
MAIN RESEARCH PROJECTS AND CONTRACTS
1. Flavin dependent systems: multitasking roles from versatile redox catalysis to cell signaling and sensing molecules. PID2022-136369NB-I00. 2023-2026. Agencia Estatal de Investigación. IP1/2: Milagros Medina/Patricia Ferreira.
2. Grupo Referencia Biología Estructural. E35_23R. 2023-2025. Gobierno de Aragón. IP1/2: Marta Martínez Júlvez/Teresa Bes Fustero.
3. Identification and synthetic applications of novel oxidative biocatalysts in industrial biotechnology. TED2021-130803B-I00. 2022-2024. Agencia Estatal de Investigación. IP1/2: Patricia Ferreira/Juan Mangas.
4. Flavoenzymes in health, disease and drug discovery. PID2019-103901GB-I00. 2020-2023. Agencia Estatal de Investigación. IP1/2: Milagros Medina/Patricia Ferreira.
5. Sistemas Enfermedades mitocondriales asociadas al mecanismo de importación y plegamiento oxidativo de proteínas en el espacio intermembrana de la mitocondria. Mecanismos moleculares y desarrollo de nuevas estrategias terapéuticas (EMPLOXPRO). LMP27_21. Dirección General de Investigación, Gobierno de Aragón. PI: P. Ferreira. 2021-2023. IP: Ferreira P
6. El flavoproteoma de Brucella: una herramienta para dianas terapéuticas y diagnósticas. LMP13_21. Dirección General de Investigación, Gobierno de Aragón. 2021-2023. IP: Medina M.
7. Paramagnetic Species in Catalysis Research. A Unified Approach Towards Heterogeneous; Homogeneous and Enzyme Catalysis (H2020 GA Number-813209). Unión Europea. Enero 2019- Diciembre 2023. IP: Dra. García I.
8. Flavoenzimas: mecanismos y dianas moleculares; patologías y aplicaciones biotecnológicas. Ministerio de Economía y Competitividad. BIO2016-75183-P. Diciembre 2016-Diciembre 2019. IP: Dra. Medina M.
PATENTS
Enzymatic Composition and Enzymatic process for the production of 2,5-furandicarboxylic acid from 5-methoxymethylfurfural using said enzymatic composition. J. Carro, E. Fernández-Fueyo, M. Alcalde, P. Ferreira, R. Ullrich, M. Hofrichter, AT. Martinez. CSIC, University of Zaragoza and Technical University of Dresden. 2017.
COLLABORATORS
From BIFI
Dr. José Antonio Ainsa.
Dr. Patricio Fernández-Silva
Dra. Raquel Moreno Loshuertos.
Dr. Victor Polo.
Dr. Adrián Velázquez-Campoy.
Dr. Javier Sancho
From other Institutions
Instituto de Nanociencia y Materiales de Aragón (INMA), Zaragoza
Dr. Jesús I. Martínez, Dra. Inés García-Rúbio, Dra Anabel Gracia Lostao.
Centro de Investigaciones Biológicas, CSIC, Madrid
Dr. Ángel Martínez.
Universitat degli Studi di Bari, Bari, Italia
Dra. Maria Barile.
Universidad de Granada, Granada
Dr. Ángel Pey.
Instituto de Recursos Naturales y Agrobiología de Salamanca, CSIC, Salamanca
Dra. Mónica Balsera.
Hospital Universitario Miguel Servet, Zaragoza.
Dr. M. Dolores Miramar, Dr. José Luis Caplabo.
Estación Experimental de Aula Dei
Dr. Manuel Becana, Dra. Inmaculada Yruela.
Centro de Investigación y Tecnología Agroalimentaria de Aragón
Dr. Pilar María Muñoz Álvaro.
Universitat Jaume I, Castellón
Dr. Vicent Moliner.
Universidad de Oviedo, Oviedo
Dr. Juan Mangas-Sanchez.
IIS Hospital 12 de Octubre, Madrid
Dr. María Morán
Universita degli Studi di Milano, Italia
Dr. Alessandro Aliverti.
Mitchell Cancer Institute, University of South Alabama, USA
Dra. Marie Migaud.
Instituto Química-Física Blas Cabrera, Madrid
Dr. José Manuel Martin.
Universita degli Studi di Torino, Turín, Italia
Dr. Salvatore Adinolfi, Dra. Sheila Sadeghi.
CONTACT
Patricia Ferreira, ferreira@unizar.es, https://orcid.org/0000-0003-4076-6118
Marta Martínez Júlvez, mmartine@unizar.es, https://orcid.org/0000-0001-9047-0046
Milagros Medina, mmedina@unizar.es, https://orcid.org/0000-0001-8743-0182
Protein misfolding and amyloid aggregation
Head of the Research Line:
Dr. Nunilo Cremades
Researchers:
Nunilo Cremades
José Daniel Camino Camino
David Polanco Irisarri
Alejandra Carrancho Arroyo
María Martínez Monge
Blanca Viguri Lamata
MAIN LINE OF RESEARCH
The phenomenon of protein misfolding and amyloid aggregation has emerged in recent years as a topic of fundamental relevance in a wide range of scientific disciplines such as physics, chemistry, biology and medicine. This explosion of interest has arisen following the recognition that approximately 50 human diseases and disorders are associated with the process of protein amyloid aggregation, some of which are among the most common and debilitating medical conditions in the modern world, including Alzheimer’s disease, Parkinson’s disease and type II diabetes.
Despite the social and economic impact of some of these diseases, the molecular origins and mechanisms of protein amyloid aggregation, as well as its associated toxicity, are unknown. In recent years, it has been proposed that aberrant liquid-liquid and liquid-solid phase separation processes of proteins may be involved in the initial events of amyloid aggregate formation in many of these diseases. The research developed in the group led by Dr. Nunilo Cremades aims to resolve these fundamental questions by combining a wide range of biophysical techniques, including single particle fluorescence methods, together with cell biology experiments. It is also intended to identify new protein targets for the development of early diagnostic tools and more effective treatments for this type of diseases.
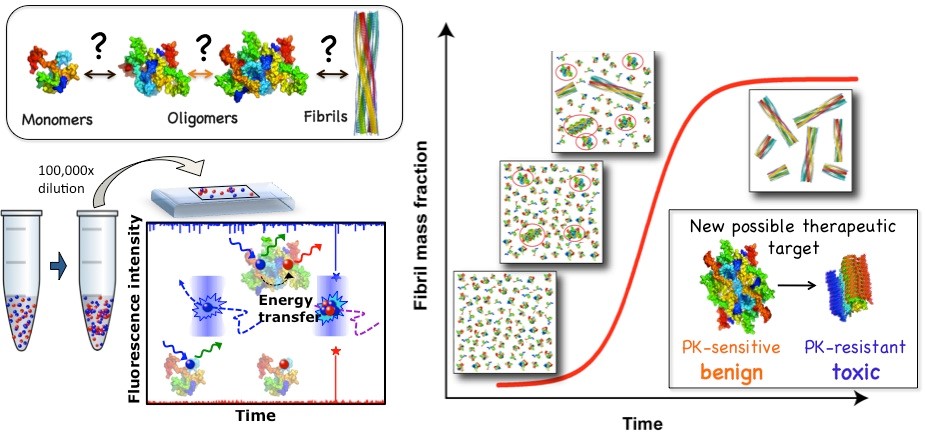 Figure 1. The development and application of single-particle fluorescence techniques has allowed us to investigate the amyloid aggregation process with an unprecedented level of detail and identify new potential therapeutic targets. Cremades N. et al. Cell 2012.
Figure 1. The development and application of single-particle fluorescence techniques has allowed us to investigate the amyloid aggregation process with an unprecedented level of detail and identify new potential therapeutic targets. Cremades N. et al. Cell 2012.
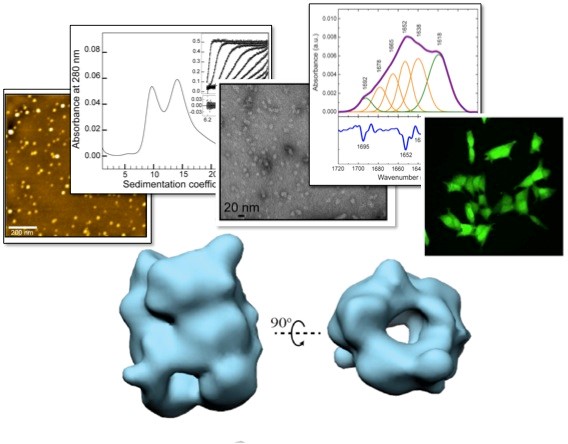 Figure 2. We have been able to isolate and purify highly stable oligomeric species of alpha-synuclein, a protein whose aggregation and deposition is associated with the development of Parkinson’s disease, and we have shown that these species are highly toxic and have general properties common with other oligomeric species generated by other amyloidogenic proteins. By combining a wide range of biophysical methods with cryo-electron microscopy image reconstruction techniques we have been able to obtain three-dimensional structural models and reveal the quaternary structural architecture of these toxic alpha-synuclein oligomers. Chen SW. et al. PNAS USA 2015; Fusco G et al. Science 2017; Cascella R et al. Nat. Commun. 2021
Figure 2. We have been able to isolate and purify highly stable oligomeric species of alpha-synuclein, a protein whose aggregation and deposition is associated with the development of Parkinson’s disease, and we have shown that these species are highly toxic and have general properties common with other oligomeric species generated by other amyloidogenic proteins. By combining a wide range of biophysical methods with cryo-electron microscopy image reconstruction techniques we have been able to obtain three-dimensional structural models and reveal the quaternary structural architecture of these toxic alpha-synuclein oligomers. Chen SW. et al. PNAS USA 2015; Fusco G et al. Science 2017; Cascella R et al. Nat. Commun. 2021
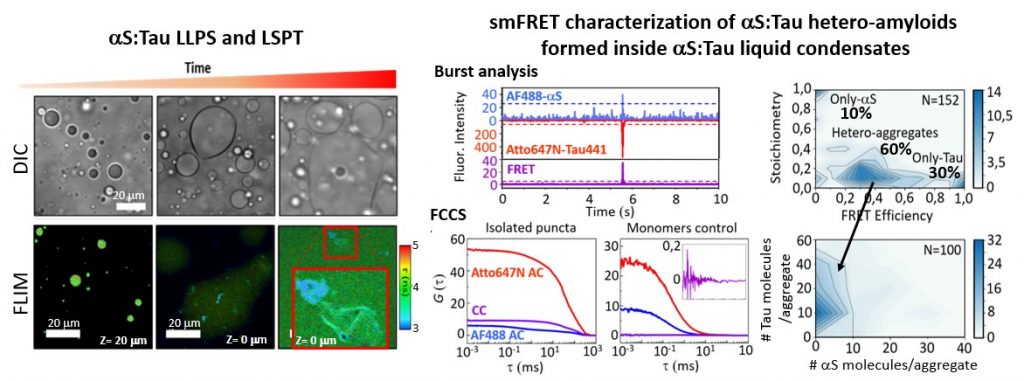 Figure 3. We have identified that alpha-synuclein (aS) and Tau proteins are capable of forming liquid condensates through a liquid-liquid phase separation (LLPS) process and that over time amyloid aggregates containing both proteins are formed inside the condensates. These processes could be involved in the formation of amyloid hetero-aggregates containing both proteins found in the brains of Parkinson’s patients. We have set up a battery of fluorescence techniques to study LLPS and LSPT (liquid-to-solid phase transitions) processes including FLIM, FCCS, smFRET. Gracia P. and Polanco D. et al. Nat. Commun. 2022
Figure 3. We have identified that alpha-synuclein (aS) and Tau proteins are capable of forming liquid condensates through a liquid-liquid phase separation (LLPS) process and that over time amyloid aggregates containing both proteins are formed inside the condensates. These processes could be involved in the formation of amyloid hetero-aggregates containing both proteins found in the brains of Parkinson’s patients. We have set up a battery of fluorescence techniques to study LLPS and LSPT (liquid-to-solid phase transitions) processes including FLIM, FCCS, smFRET. Gracia P. and Polanco D. et al. Nat. Commun. 2022
Relevant publications
1. Gracia P and Polanco D, Tarancón-Díez J, Serra I, Bracci M, Oroz J, Laurents DV, Gracía I, Cremades N*. Molecular mechanism for the synchronized electrostatic coacervation and co-aggregation of alpha-synuclein and Tau. Nat. Commun. (2022). Aug 6;13(1):4586. doi: 10.1038/s41467-022-32350-9.
2. Santos J, Gracia P, Navarro S, Peña-Díaz S, Pujols J, Cremades N*, Pallarés I*, Ventura S*. Alpha-helical peptidic scaffolds to target alpha-synuclein toxic species with nanomolar affinity. Nat. Commun. (2021) Jun 18; 12(1):3752. doi: 10.1038/s41467-021-24039-2.
3. Cascella R, Chen SW, Bigi A, Camino JD, Xu CK, Dobson CM, Chiti F, Cremades N*, Cecchi C*. The release of toxic oligomers from alpha-synuclein fibrils induces dysfunction in neuronal cells. Nat. Commun. (2021) Mar 22; 12(1):1814. doi: 10.1038/s41467-021-21937-3.
4. Camino JD, Gracia P, Chen SW, Sot J, de la Arada I, Sebastián V, Arrondo JLR, Goñi FM, Dobson CM, Cremades N*. The extent of protein hydration dictates the preference for heterogeneous or homogeneous nucleation generating either parallel or antiparallel beta-sheet alpha-synuclein aggregates. Chem. Sci. (2020) Oct 15; 11(43):11902-11914. doi: 10.1039/d0sc05297c.
5. Froula JM, Castellana-Cruz M, Anabtawi NM, Camino JD, Chen SW, Thrasher DR, Freire J, Yazdi AA, Fleming S, Dobson CM, Kumita JR, Cremades N*, Volpicelli-Daley LA*. Defining alpha-synuclein species responsible for Parkinson´s disease phenotypes in mice. J. Biol. Chem. (2019) Jul 5; 294(27):10392-10406.
6. Fusco G, Chen S., Williamson PTF, Cascella R, Perni M, Jarvis JA, Cecchi C, Vendruscolo M, Chiti F, Cremades N*, Ying L, Donson CM*, De Simone A*. Structural basis of membrane disruption and cellular toxicity by alpha-synuclein oligomers. Science (2017) Dec 15;358(6369):1440-1443.
7. Chen SW, Drakulic S, Deas E, Ouberai M, Aprile FA, Arranz R, Ness S, Roodveldt C, Guilliams T, De-Genst EJ, Klenerman D, Wood NW, Knowles TPJ, Alfonso C, Rivas G, Abramov AY, Valpuesta JM, Dobson CM*, Cremades N*. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc. Natl. Acad. Sci U.S.A. (2015) Apr 21;112(16):E1994-2003.
8. Cremades N, Cohen SI, Deas E, Abramov AY, Chen AY, Orte A, Sandal M, Clarke RW, Dunne P, Aprile FA, Bertoncini CW, Wood NW, Knowles TPJ, Dobson CM*, Klenerman D*. Direct observation of the interconversion of normal and toxic forms of alpha-synuclein. Cell (2012) 149(5): 1048-59.
Main research projects
1. GAP-101098989. IVBM-4PAP: Development of an In-Vivo Brillouin Microscope (with application to Protein Aggregation-based Pathologies). Sponsor: European Union -HORIZON-EIC-2022-PATHFINDEROPEN-01. 2023-2026. Total granted: 3.333.513 €. To the Univ. Zaragoza: 530.198,75 €. PI: N. Cremades
2. PID2022-136997NB-I00: Understanding liquid-liquid phase separation and liquid-to-solid phase transitions in amyloidogenic proteins: New paradigms in Alzheimer´s and Parkinson´s disease. Sponsor: Agencia Estatal de Investigación. 2023-2026. 250.000 €. PI: N. Cremades
3. LMP17_21: Development and validation of biomarker assays for the early diagnosis of Parkinson´s disease. Sponsor: Gobierno de Aragón. 2021-2023. 99.948,8€. PI: N. Cremades
Collaborators
Laura Volpicelli-Daley (University of Alabama, USA)
Fabrizio Chiti (University of Florence, Italy)
Janet Kumita (University of Cambridge, UK)
Douglas V. Laurents (IQFR-CSIC, Spain)
Felix Goñi (Basque Centre for Biophysics, Spain)
Arturo Muga (Basque Centre for Biophysics, Spain)
Paola Picotti (ETH Zurich, Switzerland)
Contact
Dra. Nunilo Cremades
Instituto de Biocomputación y Física de Sistemas Complejos
Universidad de Zaragoza
Mariano Esquillor S/N. Edificio I+D+i
Zaragoza 50018, Spain
Phone: +34-876555417
Email: ncc@unizar.es
Group webstie: https://sites.google.com/view/neuromol
Clinical diagnosis and drug delivery
Head of the Research Line:
Olga Abian Franco
Researchers:
Rafael Clavería, Predoctoral Student
María Arruebo, Predoctoral Student
Alberto Rodrigo, Predoctoral Student
Arturo Vinuesa, Predoctoral Student
SUMMARY OF THE RESEARCH LINE
Clinical Diagnosis
Differential Scanning Calorimetry (DSC) has recently emerged as a promising technique that provides useful information about serum/plasma interactomics (composition in proteins and metabolites, as well as their interactions). As a result of the illness, serum/plasma composition is altered and it is possible to discriminate between healthy individuals and patients with certain diseases.
A previous study performed in our group (25 healthy subjects and 60 gastric adenocarcinoma patients) showed that there were significant differences in the variables obtained from the calorimetric profiles of healthy and gastric adenocarcinoma patients and also between patients with different disease stages.
The validation and implementation of a methodology (DIGCAL) as a useful tool in diagnosing and monitoring relevant tumor pathologies (pancreatic ductal adenocarcinoma, preneoplasic pancreatic cystic lesions and stomach cancer), as well as in the isolation and identification of potential tumor biomarkers, are proposed in this project.
Calorimetric profiles of serum from healthy subjects and patients with the three types of cancer at different stages will be obtained before and after 6 months of clinical treatment. The multiparametric analysis of the thermal profiles will allow us to establish a clinical protocol for: 1/ screening a certain tumor process; 2/ classifying patients according to their tumour stage; 3/ monitoring the progression or control of the illness; 4/ identifying specific biomarkers for each disease type.
DIGCAL could reach the clinical level not only as a diagnosis tool, but also as a monitoring and tracking system of patients during the therapeutic treatment, adding value in prognostic or pharmacologic decisions. At the same time, identified potential biomarkers could be part of a user-friendly diagnosis kit for primary points of care.
Drug Delivery
Drug delivery is the method or process of administering a pharmaceutical compound to achieve a therapeutic effect in humans or animals. Drug delivery technologies modify drug release profile, absorption, distribution and elimination for the benefit of improving product efficacy and safety, as well as patient convenience and compliance. Drug release is from: diffusion, degradation, swelling, and affinity-based mechanisms. Most common routes of administration include the preferred non-invasive peroral (through the mouth), topical (skin), transmucosal (nasal, buccal/sublingual, vaginal, ocular and rectal) and inhalation routes. Many medications such as peptide and protein, antibody, vaccine and gene based drugs, in general may not be delivered using these routes because they might be susceptible to enzymatic degradation or cannot be absorbed into the systemic circulation efficiently due to molecular size and charge issues to be therapeutically effective. For this reason many protein and peptide drugs have to be delivered by injection or a nanoneedle array. For example, many immunizations are based on the delivery of protein drugs and are often done by injection.

Current efforts in the area of drug delivery include the development of targeted delivery in which the drug is only active in the target area of the body (for example, in cancerous tissues) and sustained release formulations in which the drug is released over a period of time in a controlled manner from a formulation. In order to achieve efficient targeted delivery, the designed system must avoid the host’s defense mechanisms and circulate to its intended site of action. Types of sustained release formulations include liposomes, drug loaded biodegradable microspheres and drug polymer conjugates. In this sense, Nanoparticles (NP) in Biomedicine represent a promising technology for drug transport and release. There are many possibilities for NP´s surface functionalization and so, many strategies for including drugs in them (allowing NP going mainly to their acting site) can be developed.
This research line represents a new strategy for including some antiviral compounds active against hepatitis C virus (HCV), that were previously developed in this group.
Several materials have been used:
1/ Cyclodextrins:
The chemical structure of CDs, cyclic oligosaccharides composed of α-1,4-glycosidic-linked glycosyl residues, provides them structural and physico-chemical properties that allow their use as molecular carriers.
In their hydrophobic cavity a wide range of compounds ranging from ions to proteins can be trapped. In addition, CDs exhibit low toxicity and low immunogenicity, and they have been used in the pharmaceutical field by promoting CD-drug complexes in order to improve the absorption, distribution, metabolism, excretion, and toxicity (ADMET)-related properties of a drug (eg, solubility, stability, delivery and release, membrane permeability and absorption, toxicity). Currently, more than 30 products can be found in the market based on CD complexes.
2/ Shell Cross-Linked Polymeric Micelles
Cross-linked polymeric micelles (CLPM), formed by amphiphilic block copolymers, have been successful for biomedical applications. irst, they can fulfill those general requirements for drug delivery systems: water solubility, low toxicity, to increase the stability of the drug inside the living organisms, to facilitate cellular uptake compared to free drug, and to produce its controlled release at a specific location. Second, the amphiphilic nature of the constituent polymer results in a hydrophobic core and a hydrophilic shell that allows encapsulation of both types of drug. Third, these nanoparticles offer further stability under high dilution conditions, below the critical micellar concentration, compared to other polymeric micelles.
Indeed, cross-linking avoids its disintegration in the bloodstream and the release of the drug before reaching the target cell. Particularly, fixation of the micelle structure by light-induced covalent cross-linking, mostly employing acrylate reactive groups, represents a clean and effective procedure to prepare stable polymer micelles that can hold either water-soluble and non-water soluble molecules and transport them through the bloodstream.
3/ Block Copolymers Micelles
Polymeric drug carriers are one of the current challenges of nanomedicine. Since the concept of physical drug encapsulation within polymeric aggregates was introduced, a significant number of polymer assemblies have been identified. In particular, the construction of amphiphilic block copolymer-based drug carriers is a subject of great interest and a stimulating topic of interdisciplinary research in chemistry, biology and materials science. In aqueous media, self-assembly of amphiphilic block copolymers (BCs) to minimize energetically unfavorable hydrophobic water interactions can lead to a variety of polymeric nanostructures including especially appealing spherical micelles and vesicles.
Relevant publications
1.- Polymeric micelles from block copolymers containing 2,6-diacylaminopyridine units for encapsulation of hydrophobic drugs.
Concellón A, Clavería-Gimeno E, Velázquez-Campoy A, Abian O, Piñol M, Oriol L.RSC Advances, 2016, 6, 24066–24075.
2.- Biophysical Screening for Identifying Pharmacological Chaperones and Inhibitors against Conformational and Infectious Diseases. Velazquez-Campoy A, Sancho J, Abian O, Vega S. Curr Drug Targets. 2016 Jan 31. [Epub ahead of print]
3.- Cysteine Mutational Studies Provide Insight into a Thiol-Based Redox Switch Mechanism of Metal and DNA Binding in FurA from Anabaena sp. PCC 7120. Botello-Morte L, Pellicer S, Sein-Echaluce VC, Contreras LM, Neira JL, Abian O, Velázquez-Campoy A, Peleato ML, Fillat MF, Bes MT. Antioxid Redox Signal. 2016, 24, 173-185.
4.- On the link between conformational changes, ligand binding and heat capacity. Vega S, Abian O, Velazquez-Campoy A. Biochim Biophys Acta. 2015 Oct 14. pii: S0304-4165(15)00274-3.
5.- Shell Cross-Linked Polymeric Micelles as Camptothecin Nanocarriers for anti-HCV Therapy. Jiménez-Pardo I, González-Pastor R, Lancelot A, Claveria-Gimeno R, Velázquez-Campoy A, Abian O, Ros MB, and Sierra T. Macromol. Biosci. 2015, 15, 1381–1391.
6.- Su1990 A New Technology for the Classification of Patients With Gastric Adenocarcinoma Based on Differential Scanning Calorimetry Serum Thermograms. Vega S, Garcia-Gonzalez MA, Lanas A, Velazquez-Campoy A, Abian O.*. Gastroenterology 148(4): S-569 · April 2015
7.- Rescuing compound bioactivity in a secondary cell-based screening by using γ-cyclodextrin as a molecular carrier. Clavería-Gimeno R, Vega S, Grazu V, De la Fuente JM, Lanas A, Velazquez-Campoy A, Abian O.*. International Journal of Nanomedicine 2015, 10: 2249-2259.
8.- Deconvolution Analysis for Classifying Gastric Adenocarcinoma Patients Based on Differential Scanning Calorimetry Serum Thermograms. Vega S, Garcia-Gonzalez MA, Lanas A, Velazquez-Campoy A, Abian O.*. Scientific reports, 5:7988 (2015).
9.- Ionic liquids in water: a green and simple approach to improve activity and selectivity of lipases.
Filice M, Romero O, Abian O, De las Rivas B and Palomo J.M. RSC Advances 4: 49115- 49122 (2014).
10.- A unified framework based on the binding polynomial for characterizing biological systems by isothermal titration calorimetry. Vega S., Abian O.*and Velazquez-Campoy A. Methods. 2014 pp: S1046-2023(14) 00316-8.
11.- Allosteric Inhibitors of the NS3 Protease From the Hepatitis C Virus. Abian O.*, Vega S., Sancho J., Velazquez-Campoy A. PLOSone 2013, 8 (7): 69773.
12.- NS3 protease from hepatitis C virus: Biophysical studies on an intrinsically disordered protein domain. Vega S., Neira J.L., Marcuello C., Lostao A., Abian O. * and Velazquez-Campoy A. International Journal of Molecular Sciences 2013, 14: 13282-13306.
13.- Experimental Validation of In Silico Target Predictions on Synergistic Protein Targets. Cortes-Ciriano I, Koutsoukas A, Abian O, Velazquez-Campoy A and Bender A. MedChemComm 2013, 4, 278–288.
14.- Altering the interfacial activation mechanism of a lipase by solid-phase selective chemical modification. López-Gallego F, Abian O, Guisán JM. Biochemistry. 2012 Sep 4;51(35):7028-36.
15.- Semisynthetic peptide-lipase conjugates for improved biotransformations. Romero O, Filice M, de las Rivas B, Carrasco-Lopez C, Klett J, Morreale A, Hermoso JA, Guisan JM, Abian O, Palomo JM. Chem Commun (Camb). 2012 Sep 18;48(72):9053-5.
Main research projects
Ongoing Projects:
1.- Analysis of protein/metabolites interactions in plasma serum using calorimetry: application as a quick and noninvasive diagnostic method for early detection and monitoring of tumoral digestive diseases (DIGCAL). Funding Institution: Health Institute Carlos III. From: January 2016 To: December 2018. Principal Investigator (PI): Olga Abian Franco.
2.- Validación de un nuevo método diagnóstico en suero, rápido no invasivo para detección precoz de cáncer de páncreas (PANCal). Funding Institution: Asociación Española de Gastroenterología (AEG). From: 2015 To: 2016. Principal Investigator (PI): Olga Abian Franco.
Former Projects:
3.- Adapted Nanoparticles for transport and specific release of drugs against hepatitis C virus (VHC).
Funding Institution: Health Institute Carlos III. From: 2011 To: 2013. Principal Investigator (PI): Olga Abian Franco.
4.- Implementation of in vitro e in vivo studies of anti-infectious compounds effective against Helicobacter pylori y el HCV (hepatitis C virus). Funding Institution: Health Institute Carlos III. From: 2008 To: 2011. Principal Investigator (PI): Olga Abian Franco.
Collaborators
Collaborations from BIFI
Dr. Adrián Velázquez-Campoy
Prof. Javier Sancho
Dr. Jose Luis Neira
Collaborators from other Institutions
James Graham Brown Cancer Center, University of Louisville, Louisville, KY, EEUU
PhD. Nichola Garbett
Instituto Aragonés de Ciencias de la Salud (I+CS), Zaragoza
Prof. Angel Lanas
Dra. Trinidad Serrano
Dra. Estela Solanas
Instituto de Ciencia de Materiales de Aragón (ICMA). Química Orgánica. Facultad de Ciencias.
Dra. Teresa Sierra
Prof. Luis Oriol
Prof. Milagros Piñol
Instituto de Nanociencia (INA), Universidad de Zaragoza, Zaragoza
Dr. Jesús Martínez de Lafuente
Dra. Valeria Grazú
Dra. Berta Saez
Universidad de San Jorge (USJ)
Prof. Victor López
Prof. Elisa Langa
Instituto de Catálisis y Petroleoquímica, CSIC, Madrid.
Dr. Jose Miguel Palomo
Dr. Fernando López Gallego
Dr. Jose Manuel Guisan
Universidad de Zaragoza
Dr. Jose Antonio Ainsa
Unidad de Investigación Traslacional, Hospital Universitario Miguel Servet, Zaragoza
Dra. Pilar Alfonso
Dra. Pilar Giraldo
Dr. Miguel Pocovi
Servicio de Microbiología-INIBIC. Complejo Hospitalario Universitario A Coruña, La Coruña
Dr. Francisco José Pérez-Llarena
Structural Biology of neuronal membrane receptors.
Head of the Research Line:
Beatriz Herguedas Francés
Researchers
Carlos Vega Gutiérrez (PhD student)
Irene Sánchez Valls (PhD student)
Victoria del Pilar Ribón Fuster (TFM student)
SUMMARY
AMPA type Glutamate receptors (AMPARs) are ligand-gated ion channels that mediate fast excitatory neurotransmission between neurons and play a role in neuronal plasticity. Their disfunction is associated with different disease conditions, including ALS, stroke and epilepsy. AMPARs are a diverse group of protein complexes with different kinetic, trafficking and pharmacological properties. Their diversity is achieved at different levels. The receptor core is a tetrameric ion channel composed of different combinations of four subunits (GluA1 to GluA4) that assemble as preferential heterotetramers (Herguedas et al 2013). Subunit composition is regionally and developmentally regulated (Schwenk et al 2014). There are two splicing forms for each AMPAR gene –called flip and flop- that modulate receptor kinetics, while two RNA editing sites exist in some subunits (Traynellis et al 2010). Receptor heterogeneity is further increased by the interaction with more than 30 proteins, which form both transient and stable complexes (Schwenk et al 2012). In our group we aim to explore the architecture of different AMPAR complexes using cryo-EM, getting insights into their molecular mechanism and the influence of the lipid environment in the receptor function. Besides, we aim to apply biophysical and single molecule tools to understand the dynamics of such complexes as well as to develop small molecules which target specific AMPAR complexes.
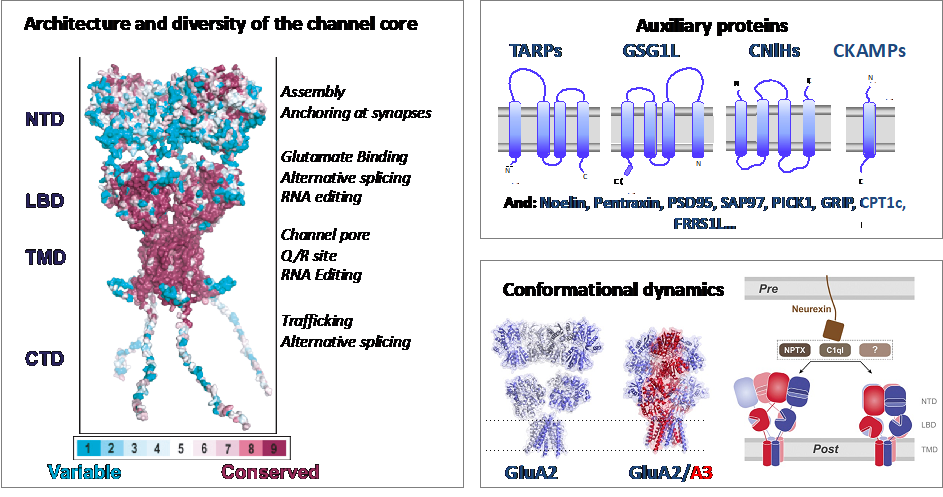 Herguedas´ group is part of the DGA group NEUROMOL, together with PIs Jose A. Carrodeguas, Nunilo Cremades, Javier García Nafría. Our focus is to study neuronal proteins -including ion channels, receptors and intrinsically disordered proteins- and their role in neuropathologies. We are also part of a the “Grupo de Acción BioF-DTE (Implementación y desarrollo de herramientas BIOFísicas en el estudio, Diagnóstico y Tratamiento de Enfermedades)” from the Campus Iberus, which aims to apply biophysical tools to develop novel strategies for the diagnosis and treatment of human diseases.
Herguedas´ group is part of the DGA group NEUROMOL, together with PIs Jose A. Carrodeguas, Nunilo Cremades, Javier García Nafría. Our focus is to study neuronal proteins -including ion channels, receptors and intrinsically disordered proteins- and their role in neuropathologies. We are also part of a the “Grupo de Acción BioF-DTE (Implementación y desarrollo de herramientas BIOFísicas en el estudio, Diagnóstico y Tratamiento de Enfermedades)” from the Campus Iberus, which aims to apply biophysical tools to develop novel strategies for the diagnosis and treatment of human diseases.
RELEVANT PUBLICATIONS
- Mechanisms underlying TARP modulation of the GluA1/2-γ8 AMPA receptor. Herguedas B, Kohegyi BK, Dohrke JN, Watson JF, Zhang D, Ho H, Shaikh SA, Lape R, Krieger JM, Greger IH. Nat Commun. 2022 Feb 8;13(1):734. doi: 10.1038/s41467-022-28404-7.
- Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP y8. Herguedas B*; Watson JF, Ho H; Cais O; García-Nafría J; Greger H*. Science, 2019 (*corresponding author)
- Druggability simulations of ionotropic glutamate receptors reveal a high-susceptibility binding site in the GluA3 AMPA receptor N-terminal domain. Lee* JY, Krieger J. *, Herguedas B. *, García-Nafría J. *, Dutta A., Shaikh S., Greger I.H., and Bahar I.. Structure, 2018. (*co-first author)
- Structure and organization of heteromeric AMPA-type glutamate receptors Herguedas B*, García-Nafría J*, Cais O, Fernández-Leiro R, Krieger J, Ho H, Greger IH.. Science. 2016. (*co-first author)
- The dynamic AMPA receptor extracellular region: a platform for synaptic protein interactionS. García-Nafría J, Herguedas B, Watson JF, Greger IH. s. J Physiol. Review. 2016.
- Structural insights into the synthesis of FMN in prokaryotic organisms. Herguedas B, Lans I, Sebastián M, Hermoso JA, Martínez-Júlvez M, Medina M. Acta Crystallogr D Biol Crystallogr. 2015.
- Mapping the interaction sites between AMPA receptors and TARPs reveals a role for the receptor N-terminal domain in channel gating. Cais O, Herguedas B, Krol K, Cull-Candy SG, Farrant M, Greger IH. Cell Reports, 2014.
- A hydrogen bond network in the active site of Anabaena ferredoxin-NADP(+) reductase modulates its catalytic efficiency. Sánchez-Azqueta A, Herguedas B, Hurtado-Guerrero R, Hervás M, Navarro JA, Martínez-Júlvez M, Medina M. BBA bioenergetics. 2014.
- Receptor heteromeric assembly-how it works and why it matters: the case of ionotropic glutamate receptors. Herguedas B, Krieger J, Greger IH. Prog Mol Biol Transl Sci. Review. 2013.
- Oligomeric state in the crystal structure of modular FAD synthetase provides insights into its sequential catalysis in prokaryotes Herguedas B, Martínez-Júlvez M, Frago S, Medina M and Hermoso JA.. J Mol Biol 2010.
- Crystallization and preliminary X-ray diffraction studies of FAD synthetase from Corynebacterium ammoniagenes. Herguedas B, Martínez-Júlvez M, Frago S, Medina M and Hermoso JA. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2009.
- Flavodoxin: a compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase. Goñi G, Herguedas B, Hervás M, Peregrina JR, De la Rosa MA, Gómez-Moreno C, Navarro JA, Hermoso JA, Martínez-Júlvez M, Medina M. BBA Bioenergetics. 2009.
- Protein motifs involved in coenzyme interaction and enzymatic efficiency in anabaena ferredoxin-NADP+ reductase. Peregrina JR*, Herguedas B*, Hermoso JA, Martínez-Júlvez M, Medina M Biochemistry. 2009. (*co-first author)
MAIN RESEARCH PROJECTS
- Structure and Dynamics of Calcium Permeable AMPA receptors. Agencia Estatal de Investigación. PID2019-106284GA-I00. (01/06/2020-31/05/2021). Principal Investigator.
- Dotación Adicional Programa Ramón y Cajal. Agencia Estatal de Investigación. RYC2018-025720-I. 01/05/2020-30/06/2025. Principal Investigator.
- AMPA Glutamate Receptors: the role of the extracellular domains in receptor assembly and allosteric regulation MRC Centenary Early Career Award. P.I. Beatriz Herguedas. 01/08/2012-30/09/2013. Principal Investigator.
COLLABORATORS
Ingo Greger (MRC Laboratory of Molecular Biology)
Javier García Nafría (Universidad de Zaragoza)
David Soto del Cerro (Universidad de Barcelona)
Raúl Estévez (Universidad de Barcelona)
Contact:
bherguedas at unizar.es
https://sites.google.com/unizar.es/herguedas-lab/
Signal transduction and membrane protein therapies
Head of the Research Line:
Javier García Nafría
Researchers:
Sandra Arroyo Urea
Ángela Carrión Antolí
Iris del Val García
Natalia Garré Ramo
Andrés Manuel González Ramirez
Javier Castillo García
SUMMARY OF THE RESEARCH LINE
Brain membrane protein complexes
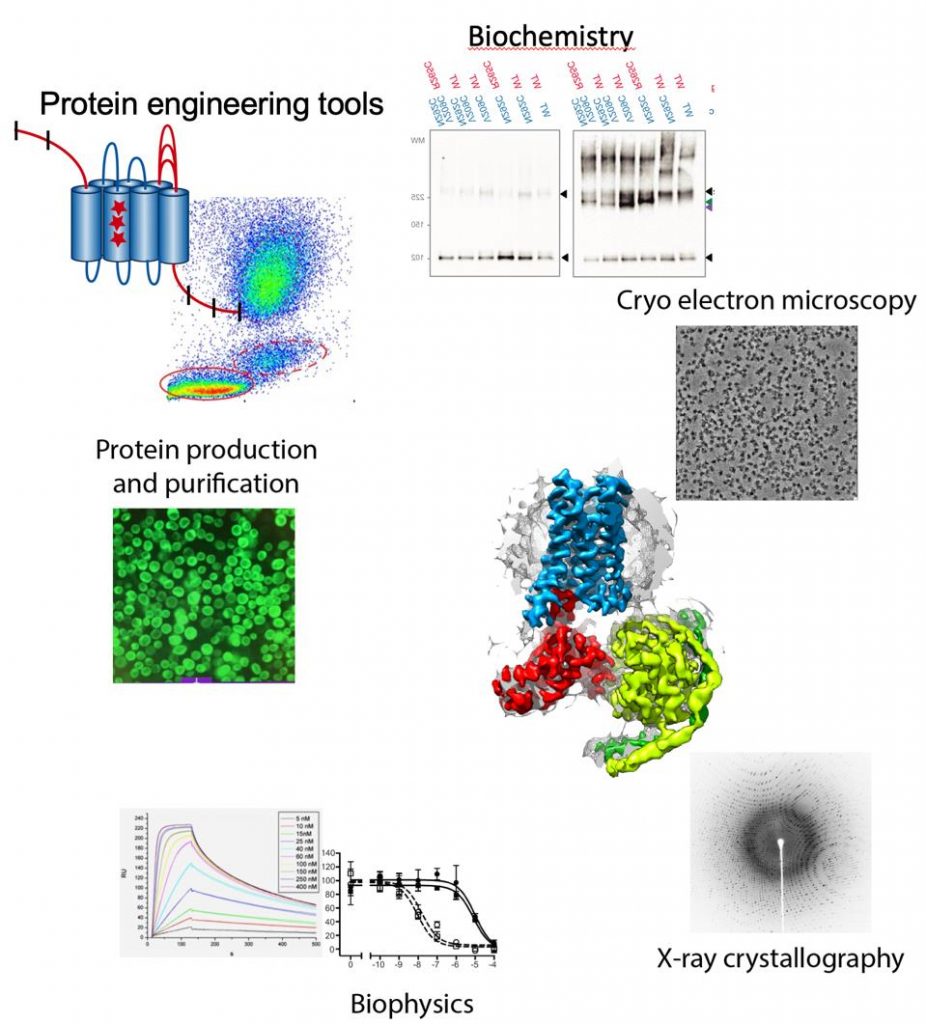
Membrane proteins represent 30% of the human genome, mediate intercellular communication and are the target for 60% of all the drugs currently used to treat disease. Membrane proteins play a especial role in the brain, where cellular communication is the basis for brain function including consciousness and memory formation. Our goal is to understand how neuronal receptors work, understanding signal detection and transduction to the intracellular millieu. We pose special emphasis on the integration of signals at the membrane by membrane protein complexes. For this purpose we use an integrated approach of structural biology techniques (Cryo-electron microscopy and X-ray crystallography), biophysics and biochemical assays. High-resolution cryo-electron microscopy currently plays a central role since we can now determine structures of membrane protein complexes that were previously intractable by X-ray crystallography.
 Protein engineering tools
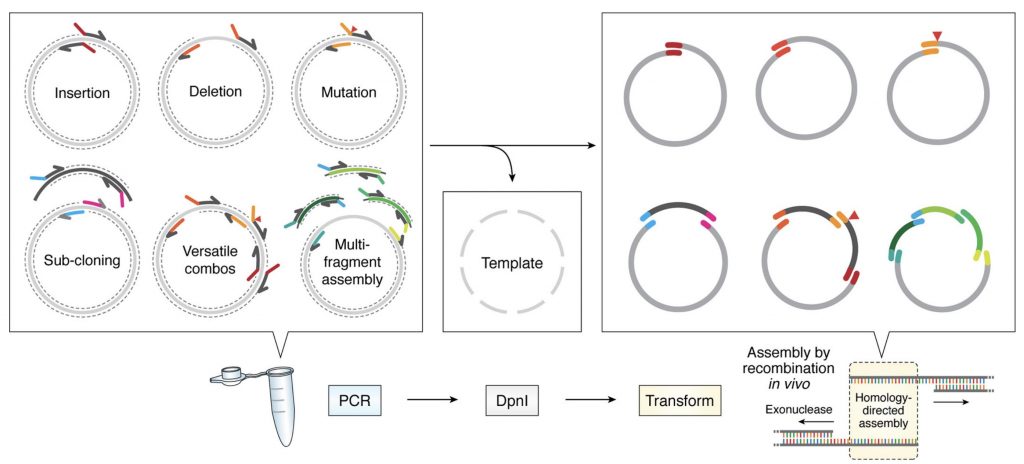
Protein engineering tools
Membrane proteins are intrinsically unstable outside the membrane environment and to overcome this problem we develop our own protein engineering tools. We have develop a molecular cloning system that simplifies all cloning protocols (García-Nafría, Sci. Rep, 2016 and Watson, JBC, 2019) , allowing to create in a cheap, quick and simple manner an array of membrane protein constructs that can be tested for expression and stability. However, this system can be applied in any biomedical research laboratory in any type of project.
Methodologies used in our research
- Molecular cloning.
- Protein engineering.
- Protein production in bacteria, insect and mammalian cells.
- Protein purification.
- Surface plasmon resonance (Biacore).
- Microscale thermophoresis.
- Macromolecular X-ray crystallography.
- Single-particle Cryo-electron microscopy.
- Biochemical cell assays (Western Blot, fluorescence, functional assays).
Relevant publications
1.New insights into GPCR coupling and dimerisation from cryo-EM structures. Gusach A, García-Nafría J, Tate CG. Curr Opin Struct Biol. 2023 Mar 22;80:102574.
2.Molecular Cloning Using In Vivo DNA Assembly. Arroyo-Urea S, Watson JF, García-Nafría J. Methods Mol Biol. 2023;2633:33-44.
3. Molecular determinants of β-arrestin coupling to formoterol-bound β1-adrenoceptor. Lee Y., Warne T., Nehme R., Pandey S., Chaturvedi M., Dwivedi-Agnihotri H., Edwards P., García-Nafría J., Leslie A., Shukla AK., Tate CG. Nature. 2020 Jun 17. doi: 10.1038/s41586-020-2419-1.
4. In vivo DNA assembly using common laboratory bacteria: a re-emerging tool to simplify molecular cloning. Jake F. Watson and García-Nafría J. Journal of Biological Chemistry. 2019 Oct 18;294(42):15271-15281.
5.Cryo-Electron Microscopy: Moving Beyond X-ray Crystal Structures for Drug receptors and Drug development.García-Nafría J.* and Tate CG*. Annual Reviews in Pharmacology and Toxicology. 2020. In press. (*corresponding author)
6. Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP γ8. Herguedas B, Watson JF, Ho H, Cais O, García-Nafría J, Greger IH. Science. 2019 Mar 14. pii: eaav9011
7. Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. García-Nafría J., Nehme R., Edwards P., Tate CG. Nature. 2018, 558 (7711). 620-62.
8. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein.García-Nafría J., Lee Y., Bai X., Carpenter B. & Tate CG. Elife. 2018, May 4;7. pii: e35946.
9. Structure and organization of heteromeric AMPA-type glutamate receptors. Herguedas B*, García-Nafria J*, Cais O, Fernandez-Leiro R, Ho H, Krieger J, Greger IH. Science. 2016, Apr 29;352(6285):aad3873.
10. IVA cloning: A single-tube universal cloning system exploiting bacterial In Vivo Assembly. García-Nafria J*, Watson JF, Greger IH. Scientific Reports. 2016, Jun 6;6:27459. (*corresponding author).
Main research projects
1. Ministerio de Ciencia e Innovación. In vitro de-orphanization of G protein-coupled receptors. MICINN. CNS2022-135877. 2023-2025.
2. Ministerio de Ciencia, Innovacion y Universidades. New modulation mechanisms of GPCRs (DopDrug): a focus on the dopamine receptors. MICINN. PID2020-113359GA-I00. 2021-2024.
3. Sosei Heptares Therapeutics. Optimization of sample production geared towards cryo-EM structure pipeline: application to orphan GPCRs. 2023-2026. Heptares Therapeutics limited.
4. Dotación adicional Ramón y Cajal. RYC2018-025731-I. 2020-2025.
Collaborators
Paula Morales, Nadine Jagerovic (IQM, CSIC, Spain)
Amy H. Newman, Alessandro Bonifazi (NIDA, NIH, USA)
Maria José Sanchez Barrena (CSIC, Madrid, Spain).
Nunilo Cremades Casasín (BIFI, Zaragoza, Spain).
CONTACT
Javier García Nafría/jgarcianafria@unizar.es
Personal group webpage: https://sites.google.com/view/signal-transduction-lab/
Enzyme modulation & Reaction Mechanisms
Head of the Research Line:
Pedro Merino
Researchers:
Manuel Pedrón (contratado pre-doctoral Gobierno de Aragón)
Sara Orta (contratado pre-doctoral Gobierno de Aragón)
Sandra Pereira (contratado pre-doctoral FPI)
Ignacio Sanz (contratado pre-doctoral FPI)
Colaboradores:
Tomas Tejero Lopez (ISQCH)
Iñaki Delso (ISQCH)
SUMMARY
SUBLINE 1: Enzyme modulation and enzymatic mechanisms
Our group focus its activity on pursuing new insights in to our understanding of health related biological processes. With a wide experience on asymmetric organic synthesis and well-equipped laboratories in the Faculty of Sciences, the group, working in the field of Chemical Biology is interested in the design and synthesis of small molecules including glycomimetics and nitrogenated compounds- acting as key modulators and/or inhibitors of target enzymes associated to specific biological functions. We are particularly interested in glycosyltransferases and transglycosylases among other enzymes. This activity is carried out by combining a series of multidisciplinary tools and techniques including in-house developed synthetic methodologies using metal- and organic catalysis, biocomputational approaches (docking and molecular dynamics) and advanced spectroscopic techniques (i.e. STD-NMR).
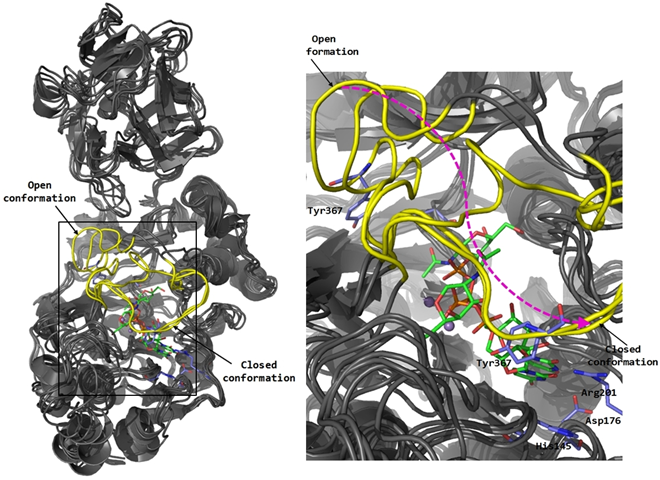 Figure 1. Glycosyltransferase GalNAc-T2: Molecular dynamics study and interactions with a designed ligand of the
Figure 1. Glycosyltransferase GalNAc-T2: Molecular dynamics study and interactions with a designed ligand of the
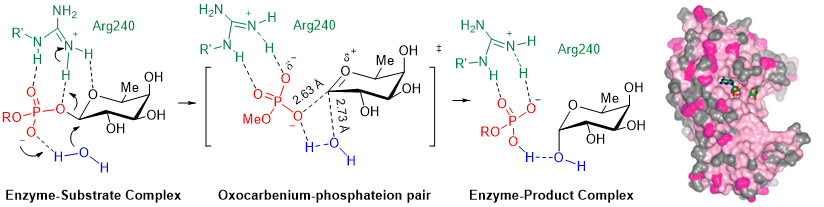 Figure 2. Glycosyltransferase POFUT1: SN1-like catalytic mechanism with formation of an intimate ion pair in the transition state
Figure 2. Glycosyltransferase POFUT1: SN1-like catalytic mechanism with formation of an intimate ion pair in the transition state
SUBLINE 2: Organic Reactions Mechanisms.
The group is also interested in studying reaction mechanisms by using QM calculations and modern topological approaches such as ELF (electron localization function) and NCI (non-covalent interactions) analyses.
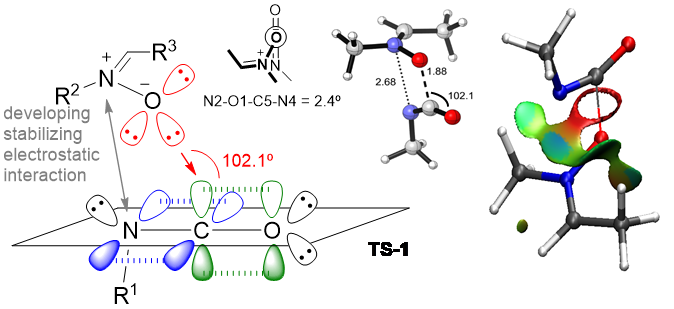 Figure 3. QM and topological studies of reaction mechanisms
Figure 3. QM and topological studies of reaction mechanisms
The studies include elucidation of catalytic reactions considering both metal-catalyzed and organocatalyzed. In particular, we are dedicated to unravel the role of chiral phosphoric acids in organocatalytic reactions in which transient carbocations are developed.
RELEVANT PUBLICATIONS
- Computational evidence of Glycosy Cations. Merino, P.; Delso, I.; Pereira, S.; Pedron, M.; Orta, S.; Tejero, T. Org. Biomol. Chem. 2021. doi: 10.1039/D0OB02373F
- Enantio- and Diastereoselective Nucleophilic Addition of N-tert-Butyl Hydrazones to Isoquinolinium Ions through Anion-Binding Catalysis. Matador, E.; Iglesias-Sigüenza, J.; Monge, D.; Merino, P.; Fernández, R.; Lassaletta, J. M. Angew. Chem. Int. Ed. 2021,133, 5096-5101. doi: 10.1002/anie.202012861
- Anomeric beta-triflate characterization enables the monitorization of glycosylation dynamics and suggests a non-canonical reinterpretation of the mechanism. Santana, A. G.; Montalvillo-Jiménez, L.; Díaz-Casado, L.; Corzana, F.; Jiménez-Osés, G.; Merino, P.; Cañada, F. J.; Jiménez-Barbero, J.; Gómez, A. M.; Asensio, J. L. J. Am. Chem. Soc. 2020, 142, 12501-12514. doi: 10.1021/jacs.0c05525
- Enantioselective Synthesis of Tropanes through Brønsted Acid-Catalyzed Pseudotransannular Desymmetrization. Rodriguez, S.; Uria, U.; Reyes, E.; Carrillo, L.; Tejero, T.; Merino, P.; Vicario, J. L. Angew. Chem. Int. Ed. 2020, 132, 6846-6850. doi: 10.1002/anie.202000650
- Dissecting the Structural and Chemical Determinants of the “Open-to-Closed” Motion in the Mannosyltransferase PimA from Mycobacteria. Unzueta, A. R.; Ghirardello, M.; Urresti, S.; Delso, I.; Giganti, D.; Anso-Miqueleiz, I.; Trastoy, B.; Comino, N.; Tersa, M.; D’Angelo, C.; Cifuente, J. O.; Marina, A.; Durana, A.; Chenal, A.; Svergun, D. I.; Alzari, P. M.; Albesa-Jové, D.; Merino, P.; Guerin, M. E. Biochemistry 2020, 59, 2934-2945. doi: 10.1021/acs.biochem.0c00376
- Enantioselective Synthesis, DFT Calculations and Preliminary Antineoplastic Activity of Dibenzo 1-Azaspiro[4.5]decanes on Drug Resistant Leukemias. Mendes, J. A.; Merino, P.; Soler, T.; Salustiano, E. J.; Costa, P. R. R.; Yus, M.; Foubelo, F.; Buarque, C. D. J. Org. Chem. 2019, 84, 2219-2233. doi: 10.1021/acs.joc.8b03203
- Sequential Metal-free Thermal 1,3-Dipolar Cycloaddition of Unactivated Azomethine Ylides. Selva, V.; Selva, E.; Merino, P.; Nájera, C.; Sansano, J. M. Org. Lett. 2018, 20, 3522-3526. doi: 10.1021/acs.orglett.8b01292
- Catalytic Enantioselective Cloke-Wilson Rearrangement. Ortega, A.; Manzano, R.; Uria, U.; Carrillo, L.; Reyes, E.; Tejero, T.; Merino, P.; Vicario, J. L. Angew. Chem. Int. Ed. 2018, 57, 8225-8229. doi: 10.1002/anie.201804614
- UDP-GlcNAc Analogs as Inhibitors of O-GlcNAc Transferase (OGT): Spectroscopic, Computational and Biological Studies. Ghirardello, M.; Perrone, D.; Chinaglia, N.; Sádaba, D.; Delso, I.; Tejero, T.; Marchesi, E.; Fogagnolo, M.; Rafie, K.; Aalten, D. M. F. v.; Merino, P. Chem. Eur. J. 2018, 24, 7264-7272. doi: 10.1002/chem.201801083
- Inhibitors against Fungal Cell Wall-remodelling Enzymes. Delso, I.; Valero-Gonzalez, J.; Gomollón-Bel, F.; Castro-López, J.; Fang, W.; Navratilova, I.; Aalten, D. M. F. v.; Tejero, T.; Merino, P.; Hurtado-Guerrero, R. ChemMedChem 2018, 13, 128-132. doi: 10.1002/cmdc.201700720
- (+)-Methyl (1R,2S)-2-{[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]methyl}-1-phenylcyclopropanecarboxylate [(+)-MR200] Derivatives as Potent and Selective Sigma Receptor Ligands: Stereochemistry and Pharmacological Properties. Amata, E.; Rescifina, A.; Prezzavento, O.; Arena, E.; Dichiara, M.; Pittalà, V.; Montilla-García, Á.; Punzo, F.; Merino, P.; Cobos, E. J.; Marrazzo, A. J. Med. Chem. 2018, 61, 372-384. doi: 10.1021/acs.jmedchem.7b01584
- Regioselectivity Change in the Organocatalytic Enantioselective (3+2) Cycloaddition with Nitrones Through Cooperative H-Bonding Catalysis/Iminium Activation. Prieto, L.; Juste-Navarro, V.; Uria, U.; Delso, I.; Reyes, E.; Tejero, T.; Carrillo, L.; Merino, P.; Vicario, J. L. Chem. Eur. J. 2017, 23, 2764-2768. doi: 10.1002@chem.201605350
- Racemic Hemiacetals as Oxygen-Centered Pronucleophiles Triggering Cascade 1,4-Addition/Michael Reaction through Dynamic Kinetic Resolution under Iminium catalysis. Reaction Development and Mechanistic Insights. Orue, A.; Uria, U.; Roca-López, D.; Delso, I.; Reyes, E.; Carrillo, L.; Merino, P.; Vicario, J. L. Chem. Sci. 2017, 8, 2904-2913. doi: 10.1039/C7SC00009J
- The small molecule luteolin inhibits N-acetyl--galactosaminyltransferases and reduces mucin-type O-glycosylation of amyloid precursor protein Liu, F.; X, K.; Xu, Z.; Rivas, M. d. l.; Li, X.; Lu, J.; Delso, I.; Merino, P.; Hurtado-Guerrero, R.; Zhang, Y. Journal of Biological Chemistry 2017, 292, 21304-21319. doi: 10.1074/jbc.M117.814202.
.
MAIN RESEARCH PROJECTS
1.- Rational Design and Stereoselective Synthesis of Glycomimetics. MINECO, 2014-2016 CTQ2013-44367-C2-1-P. 151.200 EURO. PI: Pedro Merino
2.- Rational Design of Glycomimetics Inhibitors of Glycosyltransferases. MINECO, 2017-2019 CTQ2016-76155-R. 172.800 EURO. PI: Pedro Merino
3. Estudios mecanísticos de reacciones de glicosilación y su aplicación al diseño de inhibidores de glicosiltransferasas. Ministerio de Ciencia, Innovacion y Universidades. PID2019-104090RB-100. 2020-2023. 163.350 EUROS PI: Pedro Merino Filella
COLABORATIONS
Dr. Loredana Maiuolo. Università della Calabria. Cosenza. Italy
Prof. Jose Luis Vicario. Universidad del Pais Vasco. Bilbao. Spain
Prof. Jose M. Lassaaletta. IIQ-CSIC. Sevilla. Spain.
Dr. Francesca Cardona. Universitá di Firenze. Italy
Prof. Dan van Aalten. University of Dundee. United Kingdom.
Prof. Sonsoles Martin-Santamaría. CIB-CSIC. Madrid. Spain.
Prof. Juan Luis Asensio. IQO-CSIC, Madrid. Spain
Prof. Eric Oldfield. University of Illinois, USA
Dr. Francisco Corzana. Universidad de La Rioja. Spain
Dr. Marcelo Guerin. CIC-Biogune. Bilbao. Spain
Prof. Pedro Gois. Universidade de Coimbra. Portugal.
Dr. Ramon Hurtado. Universidad de Zaragoza. BIFI. Spain
CONTACT
pmerino@unizar.es
bioorganica@unizar.es
web: http://bioorganica.unizar.es